Fluorescence microscopy is a major workhorse in life sciences, biophysics, and physical chemistry, as it allows visualization with great specificity and sensitivity. In particular, the detection of single fluorescent molecules has been used to provide information beyond ensemble averages with applications in different fields of research. For example, super-resolution methods based on the localization of single emitters have been used in the last 15 years to study biological systems with unprecedented spatial resolution in the 10-50 nm range, and even enabling the discovery of supramolecular cellular structures. On the other hand, single-molecule tracking has enabled to study individual trajectories of relevant targets that would be otherwise hidden in the average behaviour of an ensemble of unsynchronized molecules.
Most commonly, the position of a single fluorescent molecule is obtained from a fit to its image recorded in a scientific camera. However, this kind of approaches can barely surpass the 10-nm resolution barrier because of the photostability of the fluorescent dyes. In this way, the closest environment around a target (with typical size of 1-5 nm) cannot be interrogated. Alternatively, other approaches infer the molecular position from the signal registered upon excitation with a sequence of spatially shifted patterns of light. These methods, particularly when using a minimum of light intensity, have been demonstrated to be more efficient to obtain the molecular coordinates. In particular, the so-called MINFLUX technique has been proved to achieve 1-2 nm localization precision with moderate number of photons. However, the high technical complexity of MINFLUX and other related techniques has hampered its widespread application.
In our latest paper published in Light: Science & Applications, in cooperation with the group of Prof. Guillermo Acuna at the University of Fribourg (Switzerland), we present RASTMIN, a method that delivers equivalent resolution to MINFLUX, but can be implemented in standard confocal microscopes, with just two modifications (Figure 1).
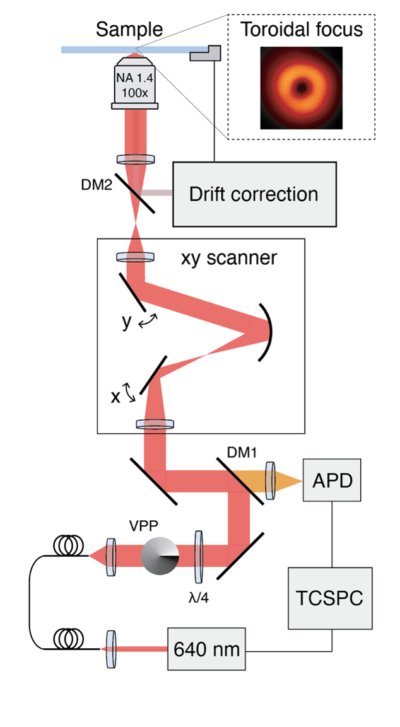
Figure 1. Schematic of a confocal microscope modified to perform RASTMIN. The two modifications are the incorporation of i) a vortex phase plate (VPP) or any other modulator to obtain a toroidal focus, and ii) a drift correction sistem to keep the sample position stable. (b)
In RASTMIN, only two main modifications to a confocal microscope need to be implemented. On the one hand, a doughnut-shaped focus has to be generated. On the other hand, the sample needs to be actively stabilized relative to the optical system. Apart from these two conditions, that are also needed in MINFLUX, no further modifications are required. For example, RASTMIN uses scanning and data acquisition routines available in any standard confocal microscope, and hence the control software already installed in any confocal setup can still be used to perform RASTMIN.
We demonstrate the performance of RASTMIN through simulations and by obtaining super-resolved images of fluorophores in a tailored DNA nanostructures (a DNA origami) achieving a localization precision of 1-2 nm (Figure 2). RASTMIN is also compatible with fluorescence lifetime imaging.
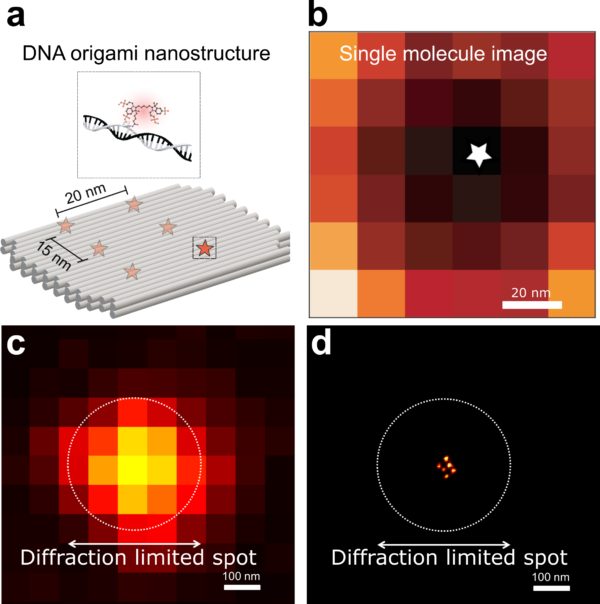
Figure 2. a. Schematic of the DNA origami used to place six fluorophores in a rectangular arrangement. b. Image of a single molecule obtained with RASTMIN. c. Diffraction limited image of the DNA origami. d. Super resolved image of the DNA origami using RASTMIN.
We anticipate that RASTMIN will pave the route to new discoveries in life sciences. So far, fluorescence imaging with nanometer-scale resolution has been limited to a reduced number of expert groups. With RASTMIN, because it can be easily implemented in existing laser-scanning microscopes, we foresee that many more groups will now access to this new regime of spatial resolution, increasing the number of biological questions addressed and discoveries.

Co-authors of the work: Prof. Fernando Stefani, Dr. Lucía Lopez, Dr. Alan Szalai, and Dr. Luciano Masullo
Read the article:
- Luciano A. Masullo, Alan M. Szalai, Lucía F. Lopez, Mauricio Pilo-Pais, Guillermo P. Acuna, and Fernando D. Stefani
“An alternative to MINFLUX that enables nanometre resolution in a confocal microscope”
News and Views about our work:
“Raster-scanning Donut simplifies MINFLUX and provides alternative implement on other scanning-based microscopes”
by Xinzhu Xu, Shu Jia & Peng Xi, Light: Science & Applications 11 (2022) 293
In the local media:
“Como lograr la máxima resolución en microscopios de uso corriente“ – Por Nora Bär



