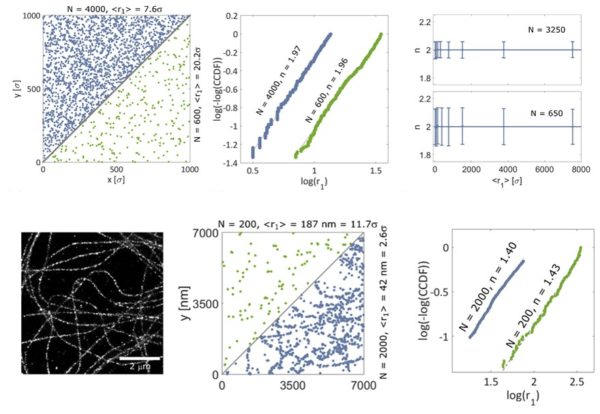
SMLM is fantastic to obtain super-resolved fluorescence images. However, it is often the case that the density of localizations is too low to obtain a clear image. Instead, sparse distributions of single-molecule positions are obtained, where any underlying molecular organization is invisible to the eye. The reason for this could be an inefficient labeling or an intrinsically low abundance of the proteins of interest. What to do then?
In our recent paper in Nanoscale, we present a method based on statistics of first-neighbor distances that reveals molecular fibrillar molecular organizations even at densities where visual inspection is useless.
This work was a collaboration with the group of Prof. Pedro Aramendía.
“Analysis of sparse molecular distributions in fibrous arrangements based on the distance to the first neighbor in single molecule localization microscopy”
Alan M Szalai, Lucía F López, Miguel Ángel Morales-Vásquez, Fernando D Stefani, Pedro F Aramendia



