DNA-origami technology is wonderful to fabricate all kinds of nanoscopic objects by self-assembly. We, in particular, use them to fabricate nanophotonic devices. That involves placing photon emitters alongside nanoparticles of different materials on well-defined nanometric geometries. DNA-origami is unique in the sense they provide great positional and stoichiometric control; we can decide where to place single molecules and single nanoparticles, one by one. This degree of control in self-assembly is fantastic, and we and others have been using DNA-origami for this reason. Now, in order to build the most efficient nanophotonic devices, positional and stoichiometric control is not enough. Orientational control is necessary because the interaction of a photon emitter with a given resonance mode is highly orientation-dependent. We have been working on this issue for some years, and in our recent paper in ACS Nano we present a method that combines super-resolution imaging with polarization-dependent excitation to determine the in-plane orientation of single molecules in DNA-origami.
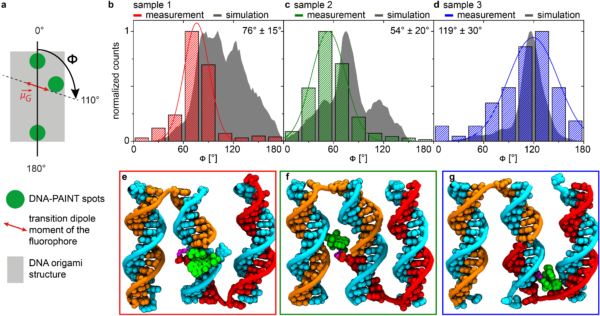
Surprisingly, we found that tuning the local environment inside the origami, different fluorescent molecules such as ATTO 647N, ATTO 643, and Cy5, take practically the same orientation. We will continue investigating this approach to control the orientation of single dyes and use it to obtain super-efficient nanophotonic devices by self-assembly.
This work is part of our ongoing, fruitful, and always fun, collaboration with the groups of Prof. Guillermo Acuna (Uni Fribourg, Switzerland) and Prof. Philip Tinnefeld (LMU Munich).
“Determining the In-Plane Orientation and Binding Mode of Single Fluorescent Dyes in DNA Origami Structures”
Kristina Hübner, Himanshu Joshi, Aleksei Aksimentiev, Fernando D. Stefani, Philip Tinnefeld, and Guillermo P. Acuna



